Transcriptomic Data Analysis
Transcriptome based investigations are the heart of biological and medical sciences researches. The advancement of next-generation sequencing technologies has given immense resolution to such investigations. It is extensively used to characterise the transcriptional activity of a cell, tissue, organism, or any biological system focusing on a subset of target genes or global gene profile under various conditions such as developmental stages, physiological variations, and in-vitro and in-vivo infection. The Next-generation sequencing technique (RNA-Seq ) explores the expression of the gene, transcript, isoforms, gene fusions, SNP in a single experiment without the limitation of prior knowledge. RNA-Seq is highly used to identify candidate genes and markers linked with particular features of interest. RNA-seq data analysis is highly demanded in academic and industrial research due to high gene and transcript level resolution, sensitivity, and accuracy. Hap Genomics provides a broad range of transcriptome data analysis services for advanced research projects and publications. Identification of the expressed genes.
1. Transcriptome assembly (Reference/de-novo) and annotation
2. Gene abundance and Differential expression analysis
3. Determination of the gene structures and splicing variants
4. Non-coding RNA analysis
5. Enrichment study of gene ontologies and pathways
6. Quantification, differential expression analysis and characterisation of known and novel microRNA
7. Target gene prediction and network/pathway analysis
8. Detection of quality variants (SNVs and InDels) and filtering of common variants in the population
9. Chip-Seq Analysis
10. Methylome-Seq Analysis
11. Metatranscriptome Analysis
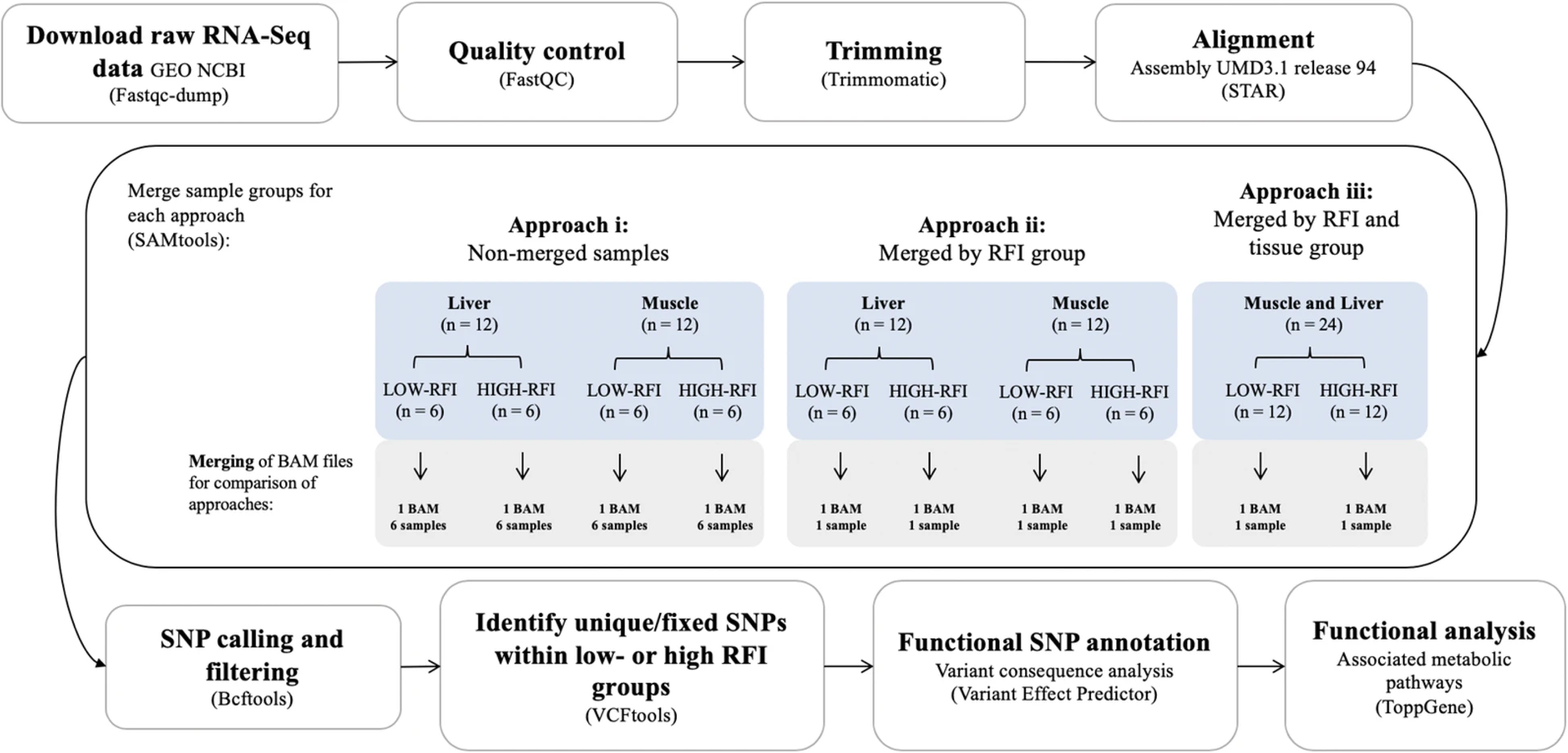
Transcriptomic Data Analysis Workflows
i) RNA-seq data analysis pipeline

ii) Functional SNP identification workflow
Results visualisation
The generated information is displayed through interactive tables and graphs that are simple to understand and export in formats suited for scientific publications.
1. Quality analysis of the sequences obtained.
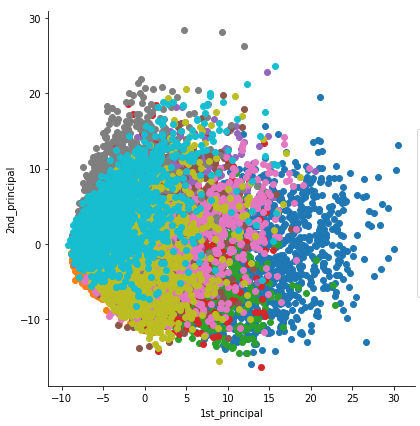
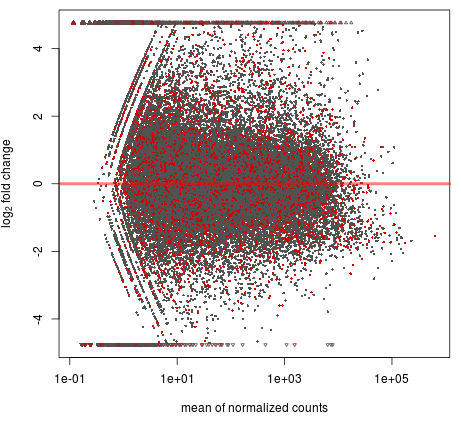
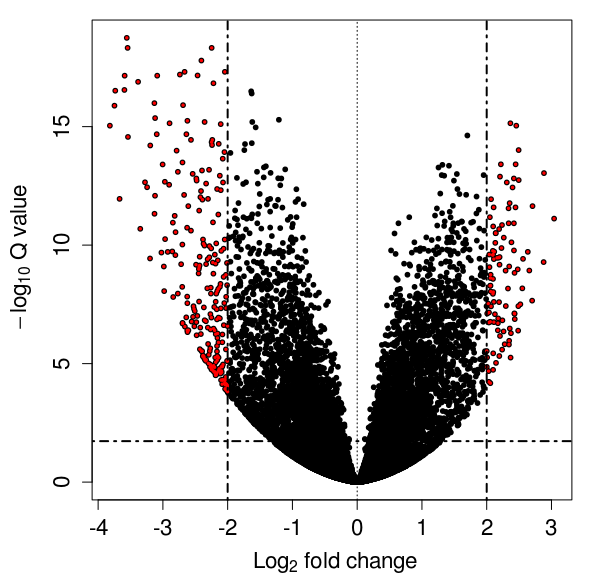
2. Sample correlation analysis and differential.
(i) PCA Plot
(ii) MA Plot/Volcano Plot
(iii) Histogram
(iv) Correlation Heatmap
3. Enrichment of Gene Ontologies paths and terms
(i) GO-MF
(ii) GO-CC
(iii) GO-BP
4. Pathway analysis: gProfiler/ WikiPathways/Panther/KEGG
Our advantages
1. Bioinformatics Consulting for Genome sequence analysis
2. Customised bioinformatics solutions
3. Publication ready data and figures
4. Manuscript drafting and support for publication
5. Qualified and experienced team
6. Delivery in time
Quick links
- Blogs
- Publications
- Top Bioinformatics Blogs
- nature research